Recently I want to write a technical post in English.
It would have been Fully modular ASP.Net assembly but I'd wait until .Net 2.1's release because it fixed a critical issue related to my project.
So I decided to write down my Data Mining course's laboratory report, as a reminder and a note.
Environment
I use Anaconda3 as my python3 env. Here is my conda's environment.yml for each experiment.
environment.segment.yml
environment.mushroom.yml
Goal
Train Decision Trees and KNN in scikit-learn against two dataset "mushroom" and "segment". Analyze theirs performance and characteristic based on the result.
Dataset "mushroom"
M. Original Data
File:
mushroom
The mushroom.dat is a space separated matrix readily for
numpy.loadtxt.
M. Decision Tree
|
1 2 3 4 5 6 7 8 |
import numpy as np from sklearn import tree f = open("mushroom.dat") data = np.loadtxt(f) features = data[...,1:23] classes = data[...,0] clf = tree.DecisionTreeClassifier() clf = clf.fit(features, classes) |
Done. Err… Let's see what we got first
|
1 2 3 4 5 |
import graphviz dot_data = tree.export_graphviz(clf, out_file=None) graph = graphviz.Source(dot_data) graph.render("mushroom") |
We got a mushroom.pdf in our working directory.
It's a graph describing the structure of the tree we built. But we can hardly exact useful information from it as there are only numbers instead of pretty names.
And the data are interpreted as continuous numeric variables, which actually is categorical variables.
The mushroom.dat we used above has already been pre-processed, so I downloaded the raw csv from https://www.kaggle.com/uciml/mushroom-classification and baked a name-number mapping using this script.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 |
csv = open("mushrooms.csv") dcsv = pd.read_csv(csv) csv.close() import re fn = open("mushroom.names") fnames = {} fdescs = {1: 'poisonous', 2: 'edible'} fncounter = 1 for line in fn: if re.match(r"\d+\.\s.*", line): fnameDesc = line.split(":") fname = fnameDesc[0].strip().split(" ")[1].strip(" ?") fnames[fncounter]=fname fdesc = map(lambda s:(s.split("=")[0].strip(), s.split("=")[1].strip()), fnameDesc[1].split(",")) for fd in fdesc: if fd[1] in dcsv[fname].unique(): col = dcsv[fname] ind = col[col == fd[1]].index[0] fdescs[int(raw_features[ind, fncounter-1])] = fd[0] fncounter+=1 fdescs = collections.OrderedDict(sorted(fdescs.items())) mapper = lambda n: fdescs[n] ndm = np.vectorize(mapper) pretty_data = ndm(data) |
|
1 2 |
fdescs = OrderedDict([(1, 'poisonous'), (2, 'edible'), (3, 'convex'), (4, 'bell'), (5, 'sunken'), (6, 'flat'), (7, 'knobbed'), (8, 'conical'), (9, 'smooth'), (10, 'scaly'), (11, 'fibrous'), (12, 'grooves'), (13, 'brown'), (14, 'yellow'), (15, 'white'), (16, 'gray'), (17, 'red'), (18, 'pink'), (19, 'buff'), (20, 'purple'), (21, 'cinnamon'), (22, 'green'), (23, 'bruises'), (24, 'no'), (25, 'pungent'), (26, 'almond'), (27, 'anise'), (28, 'none'), (29, 'foul'), (30, 'creosote'), (31, 'fishy'), (32, 'spicy'), (33, 'musty'), (34, 'free'), (35, 'attached'), (36, 'close'), (37, 'crowded'), (38, 'narrow'), (39, 'broad'), (40, 'black'), (41, 'brown'), (42, 'gray'), (43, 'pink'), (44, 'white'), (45, 'chocolate'), (46, 'purple'), (47, 'red'), (48, 'buff'), (49, 'green'), (50, 'yellow'), (51, 'orange'), (52, 'enlarging'), (53, 'tapering'), (54, 'equal'), (55, 'club'), (56, 'bulbous'), (57, 'rooted'), (58, 'missing'), (59, 'smooth'), (60, 'fibrous'), (61, 'silky'), (62, 'scaly'), (63, 'smooth'), (64, 'fibrous'), (65, 'scaly'), (66, 'silky'), (67, 'white'), (68, 'gray'), (69, 'pink'), (70, 'brown'), (71, 'buff'), (72, 'red'), (73, 'orange'), (74, 'cinnamon'), (75, 'yellow'), (76, 'white'), (77, 'pink'), (78, 'gray'), (79, 'buff'), (80, 'brown'), (81, 'red'), (82, 'yellow'), (83, 'orange'), (84, 'cinnamon'), (85, 'partial'), (86, 'white'), (87, 'brown'), (88, 'orange'), (89, 'yellow'), (90, 'one'), (91, 'two'), (92, 'none'), (93, 'pendant'), (94, 'evanescent'), (95, 'large'), (96, 'flaring'), (97, 'none'), (98, 'black'), (99, 'brown'), (100, 'purple'), (101, 'chocolate'), (102, 'white'), (103, 'green'), (104, 'orange'), (105, 'yellow'), (106, 'buff'), (107, 'scattered'), (108, 'numerous'), (109, 'abundant'), (110, 'several'), (111, 'solitary'), (112, 'clustered'), (113, 'urban'), (114, 'grasses'), (115, 'meadows'), (116, 'woods'), (117, 'paths'), (118, 'waste'), (119, 'leaves')]) fnames = {1: 'cap-shape', 2: 'cap-surface', 3: 'cap-color', 4: 'bruises', 5: 'odor', 6: 'gill-attachment', 7: 'gill-spacing', 8: 'gill-size', 9: 'gill-color', 10: 'stalk-shape', 11: 'stalk-root', 12: 'stalk-surface-above-ring', 13: 'stalk-surface-below-ring', 14: 'stalk-color-above-ring', 15: 'stalk-color-below-ring', 16: 'veil-type', 17: 'veil-color', 18: 'ring-number', 19: 'ring-type', 20: 'spore-print-color', 21: 'population', 22: 'habitat'} |
Now we have a categorical data set with meaningful values.
But before we feed them to the DecisionTreeClassifier…
Uh, DecisionTreeClassifier do not handle categorical data, "the decision trees implemented in scikit-learn uses only numerical features and these features are interpreted always as continuous numeric variables."(and there is a PR in sklearn but not merged yet).
Back to the last but two paragraph above, "The mushroom.dat we used above has already been pre-processed" — Yeah I realized that our teacher processed the data with a numeric encoding so we do not need to worry about that DecisionTreeClassifier didn't accepts the csv. And some people on SO suggests OneHotEncoder with categorical data. Will the performance differs if we apply a OneHotEncoder to the features?
M. DTC: Accuracy
We use random sampling to estimating these two pre-process method with Decision Tree Classifier’s accuracy. Here is the full python script in this step
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 |
import numpy as np import pandas as pd from sklearn import tree from sklearn.preprocessing import OneHotEncoder from sklearn.model_selection import train_test_split from sklearn.metrics import classification_report dat = open("mushroom.dat") data = np.loadtxt(dat) dat.close() raw_features = data[...,1:23] actual_classes = data[...,0] def test(test_size = 0.25): for i, rand in enumerate([21, 719, 23, 499, 153]): rf_train, rf_test, ac_train, ac_test = train_test_split(raw_features, actual_classes, test_size = test_size, random_state = rand) raw_dtc = tree.DecisionTreeClassifier(random_state = rand) raw_dtc = raw_dtc.fit(rf_train, ac_train) enc = OneHotEncoder() enc = enc.fit(raw_features) of_train = enc.transform(rf_train) of_test = enc.transform(rf_test) oh_dtc = tree.DecisionTreeClassifier(random_state = rand) oh_dtc = oh_dtc.fit(of_train, ac_train) rp = raw_dtc.predict(rf_test) op = oh_dtc.predict(of_test) print("\nRound " + str(i) + " with random_state=" + str(rand)) print("\nClassification Report of DecisionTreeClassifier with Numeric Encoding:") print(classification_report(ac_test, rp, digits=5, target_names= ["p", "e"])) print("\nClassification Report of DecisionTreeClassifier with OneHot Encoding:") print(classification_report(ac_test, op, digits=5, target_names= ["p", "e"])) |
For reproducibility, I generated five random number as random_state input. And by default train_test_split use test_size = 0.25. Let's run test() to see theirs score for the five rounds and two encoding.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 |
Round 0 with random_state=21 Classification Report of DecisionTreeClassifier with Numeric Encoding: precision recall f1-score support p 1.00000 1.00000 1.00000 974 e 1.00000 1.00000 1.00000 1057 avg / total 1.00000 1.00000 1.00000 2031 Classification Report of DecisionTreeClassifier with OneHot Encoding: precision recall f1-score support p 1.00000 1.00000 1.00000 974 e 1.00000 1.00000 1.00000 1057 avg / total 1.00000 1.00000 1.00000 2031 Round 1 with random_state=719 Classification Report of DecisionTreeClassifier with Numeric Encoding: precision recall f1-score support p 1.00000 1.00000 1.00000 985 e 1.00000 1.00000 1.00000 1046 avg / total 1.00000 1.00000 1.00000 2031 Classification Report of DecisionTreeClassifier with OneHot Encoding: precision recall f1-score support p 1.00000 1.00000 1.00000 985 e 1.00000 1.00000 1.00000 1046 avg / total 1.00000 1.00000 1.00000 2031 Round 2 with random_state=23 Classification Report of DecisionTreeClassifier with Numeric Encoding: precision recall f1-score support p 1.00000 1.00000 1.00000 972 e 1.00000 1.00000 1.00000 1059 avg / total 1.00000 1.00000 1.00000 2031 Classification Report of DecisionTreeClassifier with OneHot Encoding: precision recall f1-score support p 1.00000 1.00000 1.00000 972 e 1.00000 1.00000 1.00000 1059 avg / total 1.00000 1.00000 1.00000 2031 Round 3 with random_state=499 Classification Report of DecisionTreeClassifier with Numeric Encoding: precision recall f1-score support p 1.00000 1.00000 1.00000 931 e 1.00000 1.00000 1.00000 1100 avg / total 1.00000 1.00000 1.00000 2031 Classification Report of DecisionTreeClassifier with OneHot Encoding: precision recall f1-score support p 1.00000 1.00000 1.00000 931 e 1.00000 1.00000 1.00000 1100 avg / total 1.00000 1.00000 1.00000 2031 Round 4 with random_state=153 Classification Report of DecisionTreeClassifier with Numeric Encoding: precision recall f1-score support p 1.00000 1.00000 1.00000 939 e 1.00000 1.00000 1.00000 1092 avg / total 1.00000 1.00000 1.00000 2031 Classification Report of DecisionTreeClassifier with OneHot Encoding: precision recall f1-score support p 1.00000 1.00000 1.00000 939 e 1.00000 1.00000 1.00000 1092 avg / total 1.00000 1.00000 1.00000 2031 |
…Prefect score for all the cases. Actually even with test_size = 0.75, we still get most score eq 1.00000. And we can still get a highly accurate model with test_size = 0.95.
Not too much difference between those two encoding and OneHot may perform worse than Numeric with a small training set.
M. DTC: Visualize and Analyze
As we proved the effectiveness of our trained model, let's go back to the visualized tree. We have the name of those features now, we can get more idea from these graph.
Using random_state = 23 and entire set as training data, we got this picture:
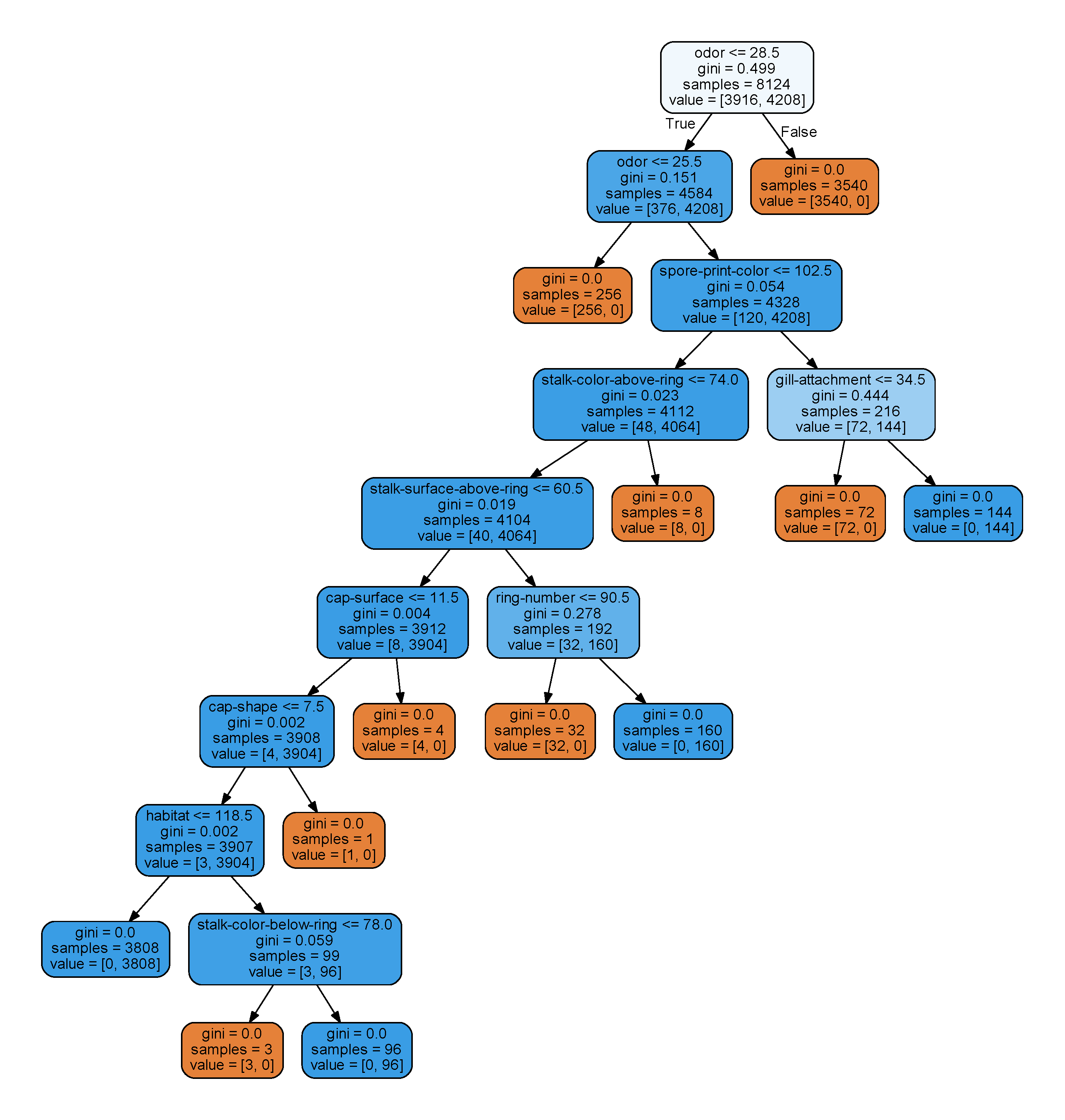
We can quickly discover the facts that odor made a significant role in telling the poisonous one from edibles as they occupy the first two nodes.
For feature odor, we have this mapping:
|
1 |
(25, 'pungent'), (26, 'almond'), (27, 'anise'), (28, 'none'), (29, 'foul'), (30, 'creosote'), (31, 'fishy'), (32, 'spicy'), (33, 'musty') |
Yup, we can say that all the mushrooms there that smell terrible are poisonous.
And for the nose-friendly mushrooms, there are small chance that you still get a poisonous one. For other features, we can also perform similar analyze.
M. K-Nearest Neighbors
For the KNN, we can reuse most code from decision tree's. I sorted out the code a little and here is the full script:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 |
import re import collections import numpy as np import pandas as pd import graphviz as gr from sklearn import tree from sklearn.tree import DecisionTreeClassifier from sklearn.neighbors import KNeighborsClassifier from sklearn.preprocessing import OneHotEncoder from sklearn.model_selection import train_test_split from sklearn.metrics import classification_report dat = open("mushroom.dat") data = np.loadtxt(dat) dat.close() csv = open("mushrooms.csv") dcsv = pd.read_csv(csv) csv.close() raw_features = data[...,1:23] actual_classes = data[...,0] fdescs = collections.OrderedDict([(1, 'poisonous'), (2, 'edible'), (3, 'convex'), (4, 'bell'), (5, 'sunken'), (6, 'flat'), (7, 'knobbed'), (8, 'conical'), (9, 'smooth'), (10, 'scaly'), (11, 'fibrous'), (12, 'grooves'), (13, 'brown'), (14, 'yellow'), (15, 'white'), (16, 'gray'), (17, 'red'), (18, 'pink'), (19, 'buff'), (20, 'purple'), (21, 'cinnamon'), (22, 'green'), (23, 'bruises'), (24, 'no'), (25, 'pungent'), (26, 'almond'), (27, 'anise'), (28, 'none'), (29, 'foul'), (30, 'creosote'), (31, 'fishy'), (32, 'spicy'), (33, 'musty'), (34, 'free'), (35, 'attached'), (36, 'close'), (37, 'crowded'), (38, 'narrow'), (39, 'broad'), (40, 'black'), (41, 'brown'), (42, 'gray'), (43, 'pink'), (44, 'white'), (45, 'chocolate'), (46, 'purple'), (47, 'red'), (48, 'buff'), (49, 'green'), (50, 'yellow'), (51, 'orange'), (52, 'enlarging'), (53, 'tapering'), (54, 'equal'), (55, 'club'), (56, 'bulbous'), (57, 'rooted'), (58, 'missing'), (59, 'smooth'), (60, 'fibrous'), (61, 'silky'), (62, 'scaly'), (63, 'smooth'), (64, 'fibrous'), (65, 'scaly'), (66, 'silky'), (67, 'white'), (68, 'gray'), (69, 'pink'), (70, 'brown'), (71, 'buff'), (72, 'red'), (73, 'orange'), (74, 'cinnamon'), (75, 'yellow'), (76, 'white'), (77, 'pink'), (78, 'gray'), (79, 'buff'), (80, 'brown'), (81, 'red'), (82, 'yellow'), (83, 'orange'), (84, 'cinnamon'), (85, 'partial'), (86, 'white'), (87, 'brown'), (88, 'orange'), (89, 'yellow'), (90, 'one'), (91, 'two'), (92, 'none'), (93, 'pendant'), (94, 'evanescent'), (95, 'large'), (96, 'flaring'), (97, 'none'), (98, 'black'), (99, 'brown'), (100, 'purple'), (101, 'chocolate'), (102, 'white'), (103, 'green'), (104, 'orange'), (105, 'yellow'), (106, 'buff'), (107, 'scattered'), (108, 'numerous'), (109, 'abundant'), (110, 'several'), (111, 'solitary'), (112, 'clustered'), (113, 'urban'), (114, 'grasses'), (115, 'meadows'), (116, 'woods'), (117, 'paths'), (118, 'waste'), (119, 'leaves')]) fnames = {1: 'cap-shape', 2: 'cap-surface', 3: 'cap-color', 4: 'bruises', 5: 'odor', 6: 'gill-attachment', 7: 'gill-spacing', 8: 'gill-size', 9: 'gill-color', 10: 'stalk-shape', 11: 'stalk-root', 12: 'stalk-surface-above-ring', 13: 'stalk-surface-below-ring', 14: 'stalk-color-above-ring', 15: 'stalk-color-below-ring', 16: 'veil-type', 17: 'veil-color', 18: 'ring-number', 19: 'ring-type', 20: 'spore-print-color', 21: 'population', 22: 'habitat'} def test_dtc(test_size = 0.75): test_clf(clf = lambda r: DecisionTreeClassifier(random_state = r), test_size = test_size) def test_knn(test_size = 0.75): test_clf(clf = lambda r: KNeighborsClassifier(), test_size = test_size) def test_clf(clf, test_size): for i, rand in enumerate([21, 719, 23, 499, 153, 348]): rf_train, rf_test, ac_train, ac_test = train_test_split(raw_features, actual_classes, test_size = test_size, random_state = rand) raw_clf = clf(rand) raw_clf = raw_clf.fit(rf_train, ac_train) enc = OneHotEncoder() enc = enc.fit(raw_features) of_train = enc.transform(rf_train) of_test = enc.transform(rf_test) oh_clf = clf(rand) oh_clf = oh_clf.fit(of_train, ac_train) rp = raw_clf.predict(rf_test) op = oh_clf.predict(of_test) print("\nRound " + str(i) + " with random_state=" + str(rand)) print("\nClassification Report of " + str(raw_clf) + " with Numeric Encoding:") print(classification_report(ac_test, rp, digits=5, target_names= ["p", "e"])) print("\nClassification Report of " + str(oh_clf) + " with OneHot Encoding:") print(classification_report(ac_test, op, digits=5, target_names= ["p", "e"])) def visualize_dtc(): clf = DecisionTreeClassifier(random_state = 23) clf = clf.fit(raw_features, actual_classes) fns = list(fnames.values()) dot_data = tree.export_graphviz(clf, out_file = None, feature_names = fns, filled = True, rounded = True) graph = gr.Source(dot_data) graph.render("mushroom_dtc") |
M. KNN: Performance and Accuracy
Runs test_knn(), we can instantly find the difference — it runs slower and is less accurate than decision tree.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 |
Round 0 with random_state=21 Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') with Numeric Encoding: precision recall f1-score support p 0.99798 0.99932 0.99865 2960 e 0.99936 0.99808 0.99872 3133 avg / total 0.99869 0.99869 0.99869 6093 Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') with OneHot Encoding: precision recall f1-score support p 1.00000 0.99865 0.99932 2960 e 0.99872 1.00000 0.99936 3133 avg / total 0.99934 0.99934 0.99934 6093 Round 1 with random_state=719 Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') with Numeric Encoding: precision recall f1-score support p 0.99629 0.99797 0.99713 2959 e 0.99808 0.99649 0.99729 3134 avg / total 0.99721 0.99721 0.99721 6093 Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') with OneHot Encoding: precision recall f1-score support p 1.00000 0.99797 0.99899 2959 e 0.99809 1.00000 0.99904 3134 avg / total 0.99902 0.99902 0.99902 6093 Round 2 with random_state=23 Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') with Numeric Encoding: precision recall f1-score support p 0.99249 0.99589 0.99419 2921 e 0.99620 0.99306 0.99463 3172 avg / total 0.99443 0.99442 0.99442 6093 Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') with OneHot Encoding: precision recall f1-score support p 0.99556 0.99795 0.99675 2921 e 0.99810 0.99590 0.99700 3172 avg / total 0.99688 0.99688 0.99688 6093 Round 3 with random_state=499 Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') with Numeric Encoding: precision recall f1-score support p 0.99793 0.99485 0.99639 2912 e 0.99530 0.99811 0.99670 3181 avg / total 0.99656 0.99655 0.99655 6093 Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') with OneHot Encoding: precision recall f1-score support p 1.00000 0.99828 0.99914 2912 e 0.99843 1.00000 0.99921 3181 avg / total 0.99918 0.99918 0.99918 6093 Round 4 with random_state=153 Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') with Numeric Encoding: precision recall f1-score support p 0.99729 0.99560 0.99644 2954 e 0.99587 0.99745 0.99666 3139 avg / total 0.99655 0.99655 0.99655 6093 Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') with OneHot Encoding: precision recall f1-score support p 1.00000 0.99729 0.99864 2954 e 0.99746 1.00000 0.99873 3139 avg / total 0.99869 0.99869 0.99869 6093 Round 5 with random_state=348 Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') with Numeric Encoding: precision recall f1-score support p 0.99451 0.99554 0.99503 2914 e 0.99591 0.99497 0.99544 3179 avg / total 0.99524 0.99524 0.99524 6093 Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') with OneHot Encoding: precision recall f1-score support p 1.00000 0.99828 0.99914 2914 e 0.99843 1.00000 0.99921 3179 avg / total 0.99918 0.99918 0.99918 6093 |
And with lower training set size, the scores drop quickly. In contrast to DTC, OneHot outperforms Numeric for most case. But those score are still good enough as most scores are higher than 0.99.
M. Further Thoughts
Why the decision tree works so well and is better than KNN approach?
We already know that odor makes a significant part in classify. With DTC, the top two root node utilizing mushroom's odor quickly distinguish most poisonous ones, but for the KNN there is no difference between one feature to the other. And the distance here didn't make too much sense here as the features are inherent categorical. And this also explains why OneHot Encoding fits well with KNN.
Here are some other things around numeric encoding that engage my attention. Although odor feature is categorical, numeric encoding coincidentally placed the poisonous smells together.
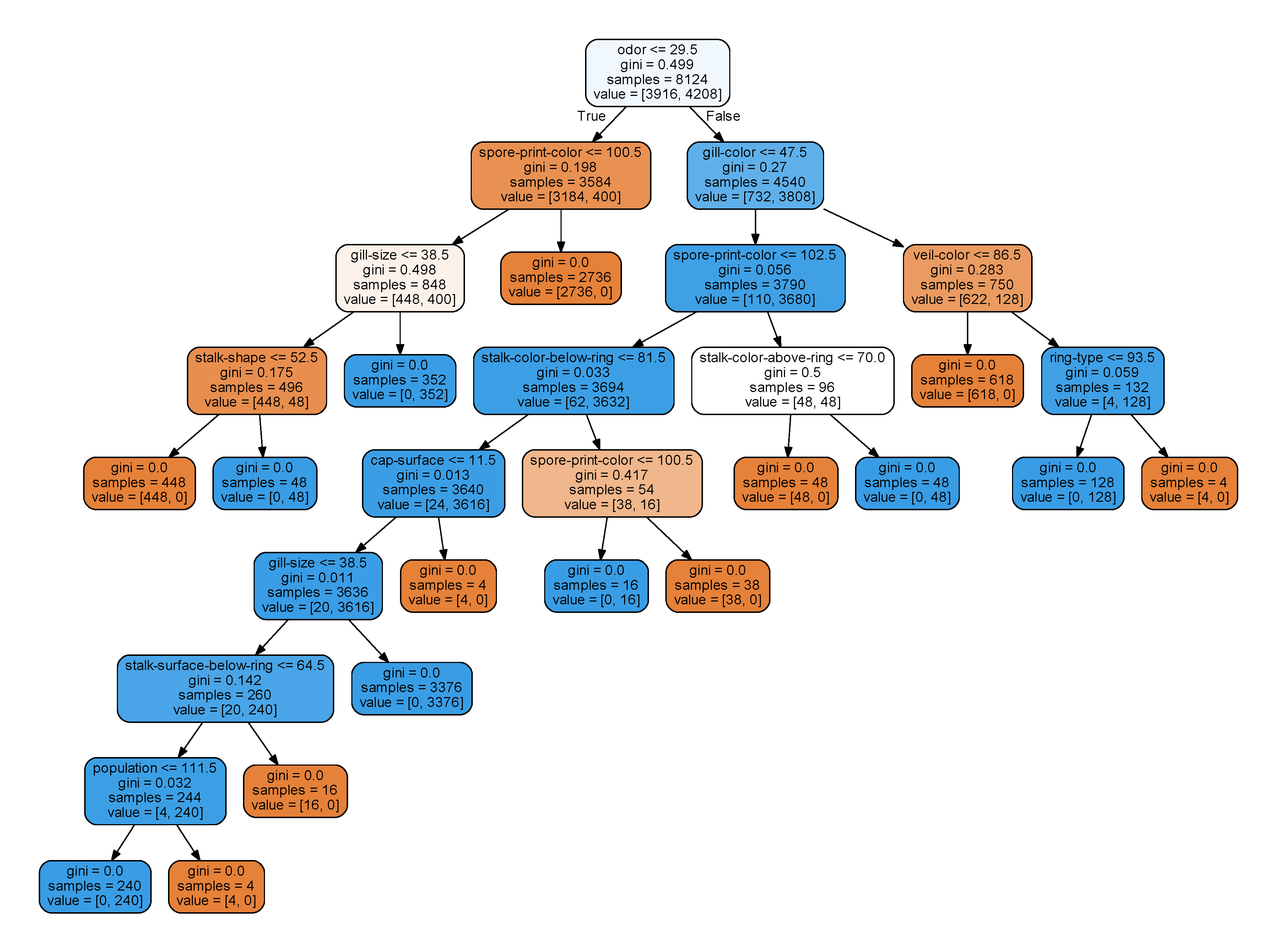
odor <= 25.5
(25, 'pungent') is poisonous
25.5 < odor <= 28.5 (26, 'almond'), (27, 'anise'), (28, 'none') most of them are edible (actually all 26s and 27s are edible) odor > 28.5
(29, 'foul'), (30, 'creosote'), (31, 'fishy'), (32, 'spicy'), (33, 'musty') all of them are poisonous
what if we mix up them a bit? I modified the data a little in mushroom.obf.dat.
(25, 'pungent') is poisonous
(26, 'foul') is poisonous
(27, 'almond') is edible
(28, 'creosote') (29, 'fishy') are poisonous
(30, 'none') is mostly edible
(31, 'spicy') is poisonous
(32, 'anise') is edible
(33, 'musty') is poisonous
Although the dataset is identical to non-obfuscated one, but it generates a very different decision tree.
The final script with all the data and reports are packed here.
mushroom.full.zip
Dataset "segment"
S. Original Data
File:
segment.zip
There are two arff-formated .txt files, "segment-train.txt" and "segment-test.txt". We can load them by scipy.io.arff and then convert them to pandas's dataframe.
S. Decision Tree
We can easily reuse the code above.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 |
import re import collections import numpy as np import pandas as pd import graphviz as gr from sklearn import tree from scipy.io import arff from sklearn.tree import DecisionTreeClassifier from sklearn.neighbors import KNeighborsClassifier from sklearn.preprocessing import OneHotEncoder from sklearn.model_selection import train_test_split from sklearn.metrics import classification_report dat = open("segment-train.txt") _, meta = arff.loadarff(dat) dat.close() dat = open("segment-train.txt") train_data = arff.loadarff(dat) dat.close() df_train = pd.DataFrame(train_data[0]) rf_train = df_train.iloc[:, :19].astype('float') ac_train = df_train.iloc[:, 19:].astype('str') dat = open("segment-test.txt") test_data = arff.loadarff(dat) dat.close() df_test = pd.DataFrame(test_data[0]) rf_test = df_test.iloc[:, :19].astype('float') ac_test = df_test.iloc[:, 19:].astype('str') def test_dtc(): test_clf(c=lambda r: DecisionTreeClassifier( random_state=r)) def test_knn(): test_clf(c=lambda r: KNeighborsClassifier()) def test_clf(c): for i, rand in enumerate([506, 286, 110, 762, 93, 418]): clf = c(rand) clf = clf.fit(rf_train, np.ravel(ac_train)) rp = clf.predict(rf_test) print("\nRound " + str(i) + " with random_state=" + str(rand)) print("\nClassification Report of " + str(clf)) print(classification_report(ac_test, rp, digits=5)) def visualize_dtc(): # raw_features = rf_test.append(rf_train, ignore_index=True) # actual_classes = ac_test.append(ac_train, ignore_index=True) raw_features = rf_train actual_classes = ac_train clf = DecisionTreeClassifier(random_state=23) clf = clf.fit(raw_features, np.ravel(actual_classes)) fns = list(raw_features) dot_data = tree.export_graphviz( clf, out_file=None, feature_names=fns, filled=True, rounded=True, class_names=meta['class'][1]) graph = gr.Source(dot_data) graph.render("segment_dtc") |
S. DTC: Performance and Accuracy
Unlike dataset "mushroom", DTC's average f1-scores are merely 0.96. That means we needs some optimization to our decision tree to achieve better performance.
At the first glance of visualized tree, I immediately spotted some clue on overfitting. Let run some test against the training set.
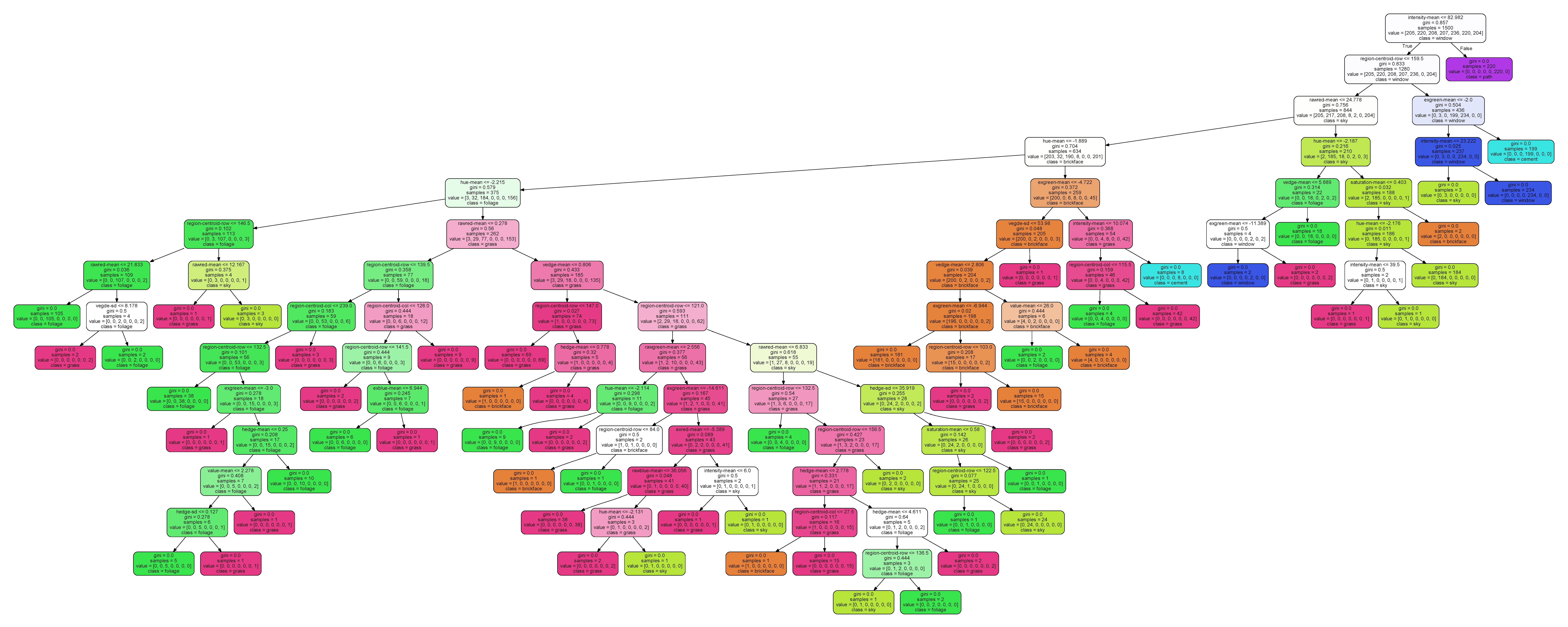
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 |
def test_dtc_train(): test_clf_train(c=lambda r: DecisionTreeClassifier( random_state=r)) def test_clf_train(c): for i, rand in enumerate([506, 286, 110, 762, 93, 418]): clf = c(rand) clf = clf.fit(rf_train, np.ravel(ac_train)) rp = clf.predict(rf_train) print("\nRound " + str(i) + " with random_state=" + str(rand)) print("\nClassification Report of " + str(clf)) print(classification_report(ac_train, rp, digits=5)) |
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 |
Round 0 with random_state=506 Classification Report of DecisionTreeClassifier(class_weight=None, criterion='gini', max_depth=None, max_features=None, max_leaf_nodes=None, min_impurity_decrease=0.0, min_impurity_split=None, min_samples_leaf=1, min_samples_split=2, min_weight_fraction_leaf=0.0, presort=False, random_state=506, splitter='best') precision recall f1-score support b'brickface' 1.00000 1.00000 1.00000 205 b'cement' 1.00000 1.00000 1.00000 220 b'foliage' 1.00000 1.00000 1.00000 208 b'grass' 1.00000 1.00000 1.00000 207 b'path' 1.00000 1.00000 1.00000 236 b'sky' 1.00000 1.00000 1.00000 220 b'window' 1.00000 1.00000 1.00000 204 avg / total 1.00000 1.00000 1.00000 1500 Round 1 with random_state=286 Classification Report of DecisionTreeClassifier(class_weight=None, criterion='gini', max_depth=None, max_features=None, max_leaf_nodes=None, min_impurity_decrease=0.0, min_impurity_split=None, min_samples_leaf=1, min_samples_split=2, min_weight_fraction_leaf=0.0, presort=False, random_state=286, splitter='best') precision recall f1-score support b'brickface' 1.00000 1.00000 1.00000 205 b'cement' 1.00000 1.00000 1.00000 220 b'foliage' 1.00000 1.00000 1.00000 208 b'grass' 1.00000 1.00000 1.00000 207 b'path' 1.00000 1.00000 1.00000 236 b'sky' 1.00000 1.00000 1.00000 220 b'window' 1.00000 1.00000 1.00000 204 avg / total 1.00000 1.00000 1.00000 1500 ...... |
Yep, full marks. We got an slightly over-fitting tree. Let's follow the tips from http://scikit-learn.org/stable/modules/tree.html#tips-on-practical-use and seek better performance.
|
1 2 3 |
def test_dtc(min_samples_split=3, max_depth=10, criterion='gini', splitter='best'): test_clf(c=lambda r: DecisionTreeClassifier( random_state=r, min_samples_split=min_samples_split, max_depth=max_depth, criterion=criterion, splitter=splitter)) |
After some rounds of tests, I got a better score with (min_samples_split=10, max_depth=9, criterion='entropy', splitter='best')
S. DTC: Visualize
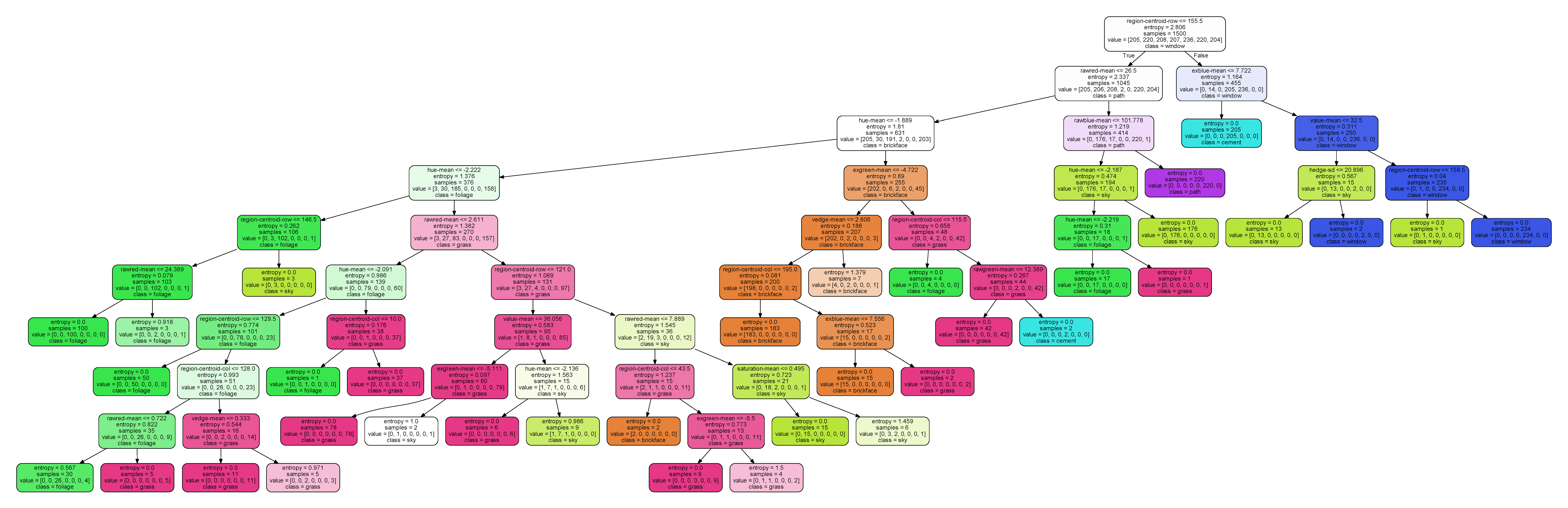
S. K-Nearest Neighbors
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 |
from sklearn.preprocessing import normalize nf_train = normalize(rf_train) nf_test = normalize(rf_test) def test_knn(n_neighbors=5): test_clf(c=lambda r: KNeighborsClassifier( n_neighbors=n_neighbors), rnd=[1]) def test_knn_n(n_neighbors=5): test_clf(c=lambda r: KNeighborsClassifier( n_neighbors=n_neighbors), rf_train=nf_train, rf_test=nf_test, rnd=[1]) def test_clf(c, rf_train=rf_train, rf_test=rf_test, rnd=[506, 286, 110, 762, 93, 418]): for i, rand in enumerate(rnd): clf = c(rand) clf = clf.fit(rf_train, np.ravel(ac_train)) rp = clf.predict(rf_test) if len(rnd) > 1: print("\nRound " + str(i) + " with random_state=" + str(rand)) print("\nClassification Report of " + str(clf)) print(classification_report(ac_test, rp, digits=5)) |
S. KNN: Performance and Accuracy
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 |
>>> test_knn() Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') precision recall f1-score support b'brickface' 0.88806 0.95200 0.91892 125 b'cement' 0.91667 0.90000 0.90826 110 b'foliage' 0.90083 0.89344 0.89712 122 b'grass' 1.00000 0.99187 0.99592 123 b'path' 0.94949 1.00000 0.97409 94 b'sky' 1.00000 1.00000 1.00000 110 b'window' 0.86207 0.79365 0.82645 126 avg / total 0.92915 0.92963 0.92891 810 >>> test_knn(1) Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=1, p=2, weights='uniform') precision recall f1-score support b'brickface' 0.95238 0.96000 0.95618 125 b'cement' 0.92857 0.94545 0.93694 110 b'foliage' 0.93333 0.91803 0.92562 122 b'grass' 1.00000 0.99187 0.99592 123 b'path' 1.00000 1.00000 1.00000 94 b'sky' 1.00000 1.00000 1.00000 110 b'window' 0.86508 0.86508 0.86508 126 avg / total 0.95192 0.95185 0.95186 810 >>> test_knn_n(1) Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=1, p=2, weights='uniform') precision recall f1-score support b'brickface' 0.96063 0.97600 0.96825 125 b'cement' 0.85321 0.84545 0.84932 110 b'foliage' 0.91818 0.82787 0.87069 122 b'grass' 1.00000 0.96748 0.98347 123 b'path' 0.91000 0.96809 0.93814 94 b'sky' 0.96491 1.00000 0.98214 110 b'window' 0.82443 0.85714 0.84047 126 avg / total 0.91915 0.91852 0.91823 810 >>> test_knn_n() Classification Report of KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski', metric_params=None, n_jobs=1, n_neighbors=5, p=2, weights='uniform') precision recall f1-score support b'brickface' 0.96721 0.94400 0.95547 125 b'cement' 0.77966 0.83636 0.80702 110 b'foliage' 0.83607 0.83607 0.83607 122 b'grass' 0.99180 0.98374 0.98776 123 b'path' 0.86792 0.97872 0.92000 94 b'sky' 0.98214 1.00000 0.99099 110 b'window' 0.87037 0.74603 0.80342 126 avg / total 0.90116 0.90000 0.89928 810 |
I don't why normalizing the data leads to worse result…And the best k for this dataset is 1…
And the final script here segment.full.zip
![]() [Dr.Lib] Data Mining: KNN and Decision Tree by Liqueur Librazy is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
[Dr.Lib] Data Mining: KNN and Decision Tree by Liqueur Librazy is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.